Abstract
In recent years, brain hemorrhage has rapidly increased and poses a significant danger to life. Automatic detection and classification of brain hemorrhage are crucial for doctors. This paper presents a new approach using convolutional neural networks and Hounsfield Unit values for analyzing brain hemorrhage from CT/MRI images. The method involves two stages: classification of brain hemorrhage using convolutional neural networks, and detection of hemorrhage areas and determination of hemorrhage time based on Hounsfield Unit values. This method is effective for doctors in diagnosing the location, time, and severity of brain hemorrhage for prompt treatment.
Introduction
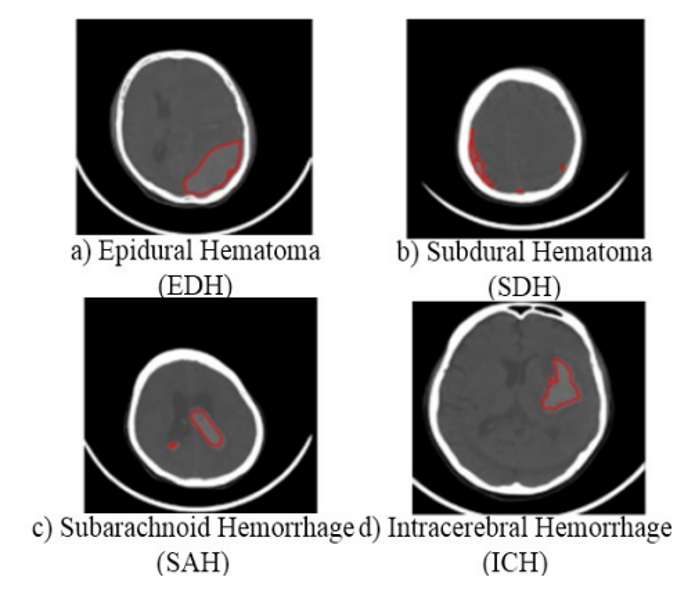
Stroke is one of the most dangerous diseases and the third leading cause of death for humans. According to the Vietnam National Heart Association, in 2005, 16 million people suffered a stroke for the first time, and 5.7 million died from it. Stroke is divided into two types: ischemic stroke (20%) and hemorrhagic stroke (80%). Brain hemorrhage can be deadly, requiring quick and accurate medical diagnosis. The brain's complex structure makes symptom recognition difficult, leading to potential errors in CT/MRI image analysis. Stressful working conditions for doctors further complicate accurate diagnosis. Therefore, automatic diagnosis systems integrated with CT/MRI systems are essential to reduce errors and shorten diagnosis time. Recent studies suggest using Convolution Neural Network (CNN), a deep learning model that improves image classification accuracy without the need for feature extraction. Companies like Google and Facebook use CNN for image and speech processing. CNN's application in medical image classification, such as detecting intracranial hemorrhage, has shown promising results. However, limitations include the requirement for large training datasets, powerful GPUs, and challenges in pinpointing the exact location or duration of hemorrhages. To address these issues, a new approach using CNN is proposed for automatic classification of brain hemorrhage, focusing on epidural hematoma (EDH), subdural hematoma (SDH), subarachnoid hemorrhage (SAH), and intracerebral hemorrhage (ICH). The Hounsfield Unit (HU) is used to determine the area and timing of hemorrhage for effective treatment.
Proposed Method
The system consists of two phases: the first phase classifies MRI images to label hemorrhage types; the second phase segments images using HU values to clarify the hemorrhagic region and duration. Input images are processed through a CNN-based classification followed by HU-based image segmentation for the damage area and duration.
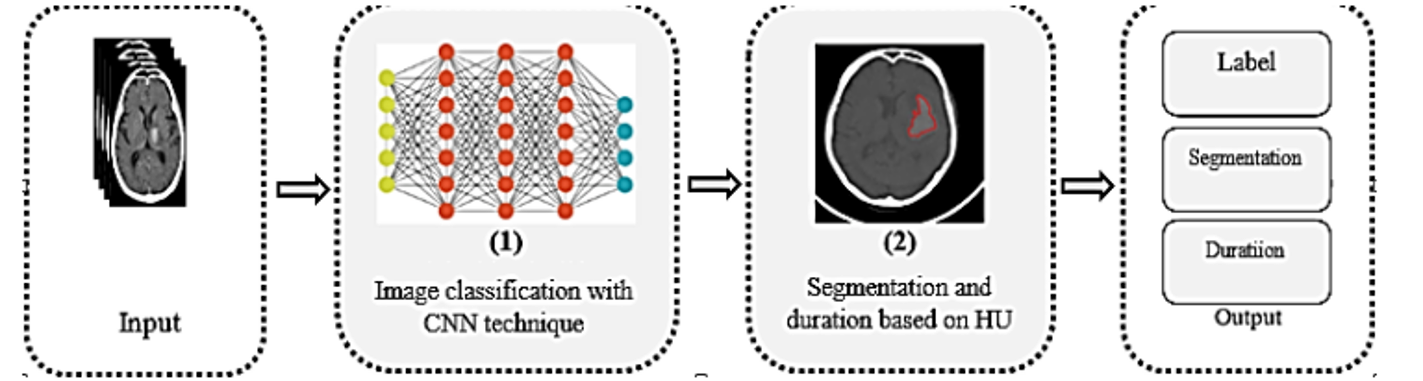
A. Phase 1: Image classification with CNN technique
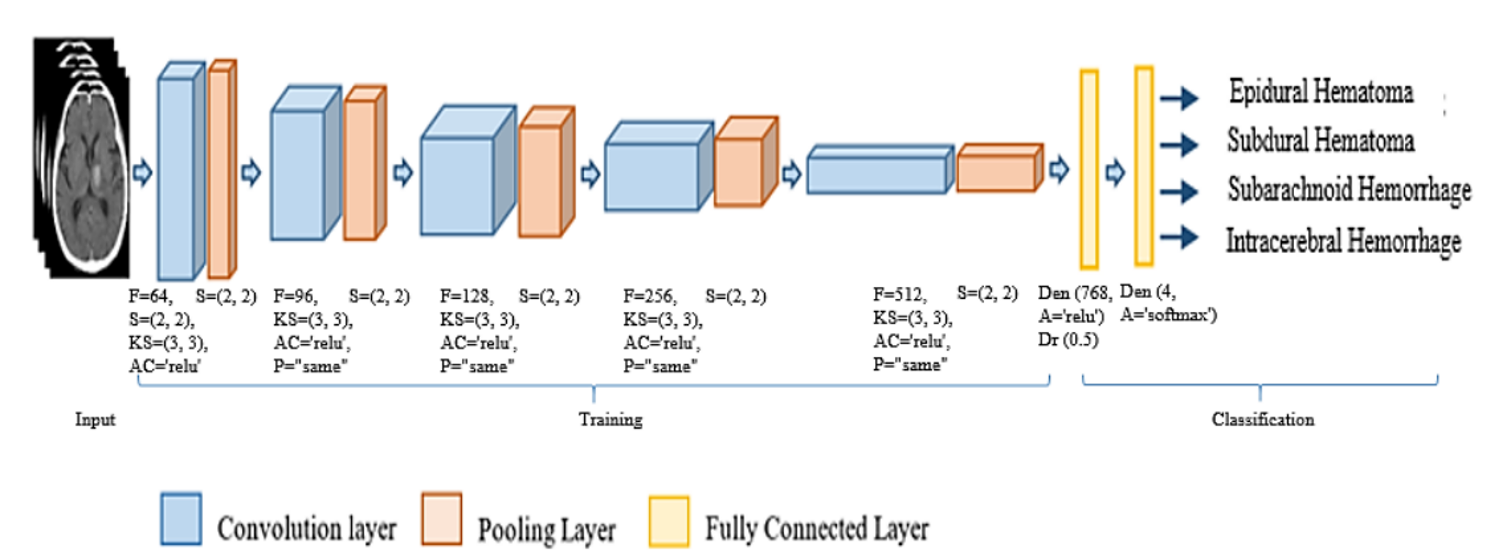
The CNN model of this phase consists of convolution layers, pooling layers, and fully connected layers. This phase includes two steps: Step 1, training image features; Step 2, classifying hemorrhage based on the learning model. In step 1, we use 5 convolution layers, 5 pooling layers, and 2 fully connected layers. For the convolution layer, we use parameters: number of filters F = 64, strides S = (2, 2), kernel size KS = (3, 3), activation function AC = ‘relu’, and padding P = ‘same’. For pooling layers, we use a filter S of size (2x2). We use two dense layers with parameters: number of output nodes and activation = ‘relu’. Dropout, with Dr = 0.5, is used to reduce overfitting.
Feature training
a) Convolution layer: We use five convolution layers for high accuracy with shorter training times. ReLU is used for nonlinear transformation.
b) Pooling layer: Max Pooling is used with a (2x2) filter to reduce input space size and computational cost, while highlighting important features.
Classification
The fully connected layer includes the dense layer and dropout layer.
a) Dense layer: Connects layers in a transitional fashion, enhancing features, reducing gradient, and reusing features. Functions include ReLU and Softmax for classification.
b) Dropout layer: Randomly skips data from hidden layers to reduce interdependence between neurons.
The result is labeled classification of images into four types of cerebral hemorrhage: epidural hematoma, subdural hematoma, subarachnoid hemorrhage, and intracerebral hemorrhage.
B. Phase 2: Segmentation and hemorrhage duration based on HU
Phase 2 calculates HU values for locating and determining the duration of cerebral hemorrhage, providing additional information such as hemorrhage region and bleeding time for timely treatment.
Results
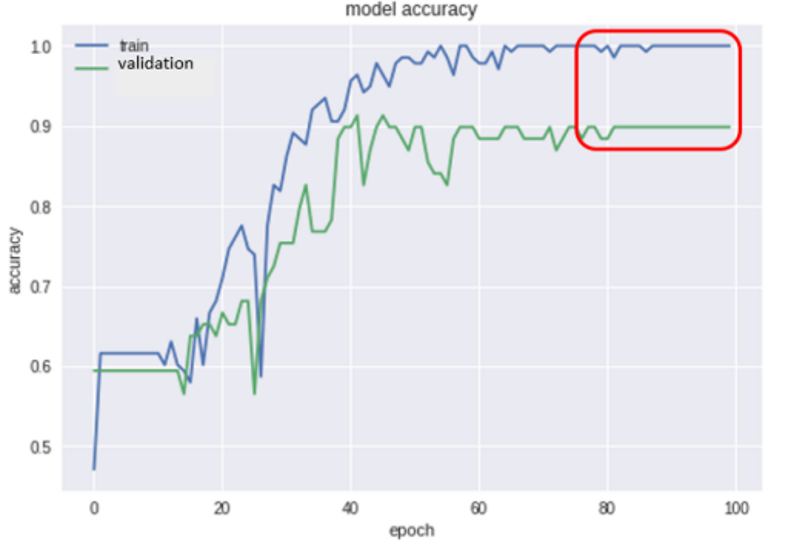
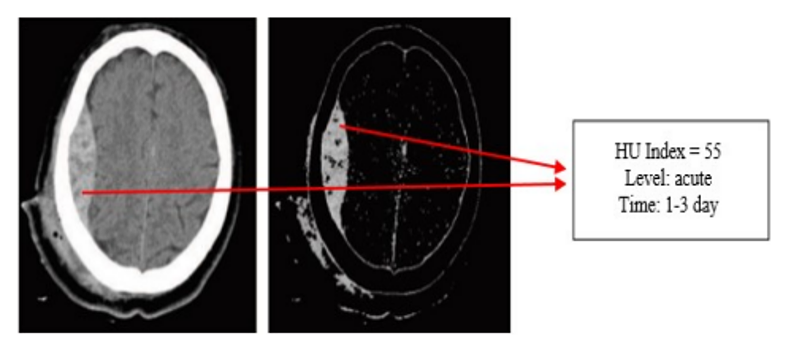
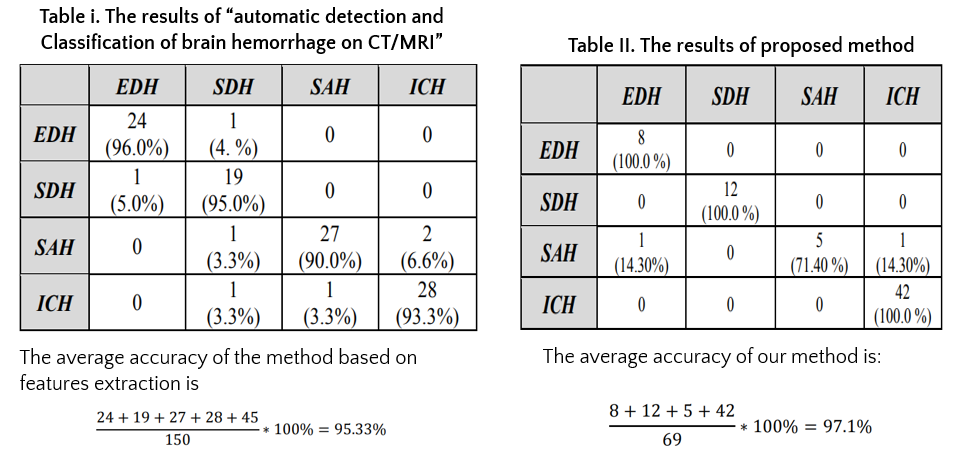
In addition, we compared the proposed method to the method based on features extraction
Conclusions
In this paper, we propose a new approach for the detection and classification of brain hemorrhage based on convolutional neural network techniques and HU values, which are commonly used by specialists in detecting brain hemorrhage. Our proposed approach not only classifies brain hemorrhage but also determines the accurate region and duration of hemorrhaging to effectively support doctors in treatment. Experimental results of the model show that the classification accuracy was 97.1%, and the accurate location of brain hemorrhage was determined. However, we have some limitations, such as a limited number of data sets and the lack of comparison with other advanced classification methods. Additionally, we experimented with only one of the four types of brain hemorrhage from single-bleeding patients. These issues will be addressed in our future research.
References
- Nguyễn Văn Chi, “Cập nhật về chẩn đoán và xử trí đột quỵ não cấp”, Hội nghị Tim mạch Toàn quốc, 2016.
- Lectures of Stanford University, “Introduction to Convolutional Neural Networks”, 2018.
- S. González-Villà, A. Oliver, S. Valverde, L. Wang, R. Zwiggelaar, X. Lladó, “A review on brain structures segmentation in magnetic resonance imaging”, Artificial Intelligence in Medicine 73, 2016, pp. 45–69.
- J. Mitra, P. Bourgeat, J. Fripp, S. Ghose, S. Rose, O. Salvado, A. Connelly, B. Campbell, S. Palmer, G. Sharma, et al., “Lesion segmentation from multimodal CT/MRI using random forest following ischemic stroke”, Neuroimage 98, 2014, pp. 324–335.
- E. Roura, A. Oliver, M. Cabezas, S. Valverde, D. Pareto, J. C. Vilanova, L. Ramió-Torrentà, À. Rovira, X. Lladó, “A toolbox for multiple sclerosis lesion segmentation”, Neuroradiology 57 (10), 2015, pp. 1031–1043.
- K.-k. Shen, J. Fripp, F. Mériaudeau, G. Chételat, O. Salvado, P. Bourgeat, A. D. N. Initiative, et al., “Detecting global and local hippocampal shape changes in Alzheimer’s disease using statistical shape models”, Neuroimage 59 (3), 2012, pp. 2155–2166.
