Abstract
In this study, I propose a novel approach of automatic classification of common liver lesions based on deep learning techniques and the variation in Hounsfield Unit intensity on CT images in the pre- and post-contrast periods. The method I propose is to rely on the variation in the intensity of the Hounsfield index on CT images in the pre- and post-contrast periods, to accurately identify the lesion area on the liver to support data labeling. Deep learning techniques such as Faster R-CNN, R-FCN and Mask R-CNN with characteristic extraction networks such as ResNet 101, Inception-V2, Inception-ResNet-V2, are used to automatically detect and classify liver lesions. The experimental results show that the proposed method with the Faster R-CNN ResNet-101 network model achieves a higher accuracy than the other network model with the measured mAP of 97.4%. This method effectively supports doctors to accurately locate and classify lesions to have timely treatment directions for patients.
Introduction
Polluted environments, chemical residues in food, and poor living habits have increased cancer rates globally, with liver cancer being particularly prevalent. According to GLOBOCAN, in 2020 there were about 905,677 new cases and 830,180 deaths from liver cancer, ranking it 7th among cancers worldwide (Sung et al., 2021). In Vietnam, liver cancer had 26,418 new cases in 2020, making it the most common cancer in men and the fifth in women (Tran Le and Viet Dao, 2020). This highlights the urgent need for early detection and classification of liver lesions. CT imaging is a key tool in diagnosing and monitoring liver lesions (Taylor and Ros, 1998). However, current detection largely relies on doctors' expertise, which can be challenging for subtle lesions. Advances in artificial intelligence, particularly deep learning, offer promising improvements in diagnostic accuracy and efficiency. Despite the reliance on the Hounsfield Unit (HU) index for detecting abnormalities, previous studies have often overlooked its use. In response, my research, titled "Detection and Classification of Common Liver Lesions on CT Images Based on the Hounsfield Index and Deep Learning Techniques," aims to enhance early liver pathology detection and support timely treatment solutions.
Proposed Method
In this study, I propose a three-stage generalized model consisting of: (1) preprocessing to prepare input data for the deep learning model, (2) feature extraction and training, and (3) vulnerability detection and classification. Figure 1 illustrates this model, detailing the three stages: data preprocessing, feature extraction, and detection and classification.
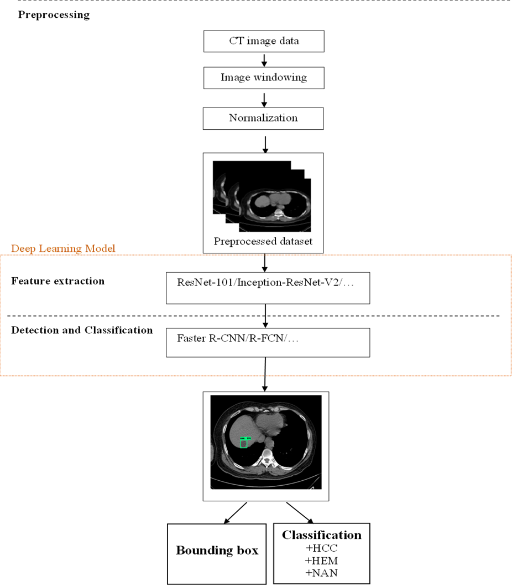
The difference of this study is that it is processed on the input data of 4 CT image sets before and after contrast injection. The input image of the study is a CT image set in Dicom format, so in order to obtain a suitable image set for the training and testing process, it is necessary to build and process the data set as shown in Figure 2.
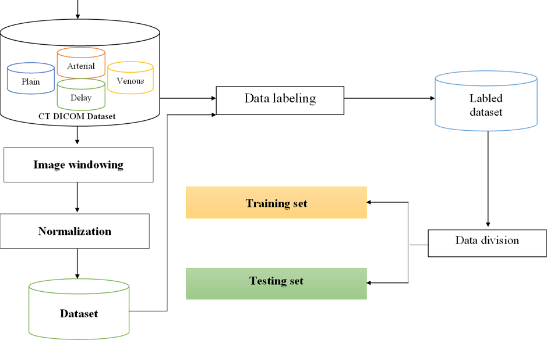
After preprocessing and labeling the dataset, export the files needed for the training process, most importantly the record.record and labelmap.pbtxt sets. When train photos are brought in for training, they will be extracted through the feature extraction networks to extract the corresponding features.
Results
In order for the experimental results to be objective and complete, all three types of lesions are HCC, HEM, and NAN. I have selected 3 images with small lesions or faint lesions that are difficult to notice and included in 8 experimental models to evaluate and present in this section.
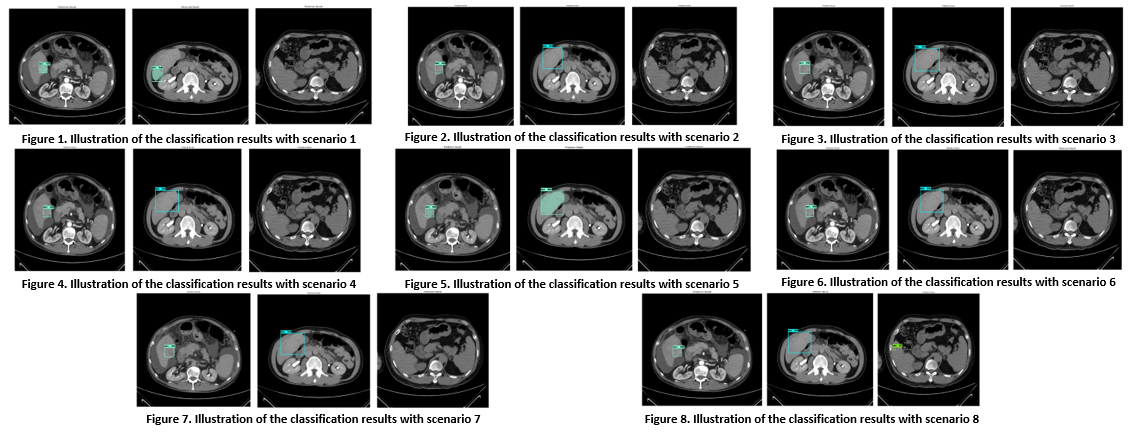
Through the experimental process for the scenarios that the topic proposes. It can be generally assessed that, for medium and large lesions, it is easy to see; when this image is included in the proposed experimental models, it can be seen that the detection and classification results are quite accurate. However, for some unknown lesions, the intensity of which is similar to liver tissue and the size of the lesion is small, the results are still inaccurate; sometimes the lesion is not visible or the label is wrong. However, with the idea of improving the classification model as proposed in this study, it has helped to limit misclassification.
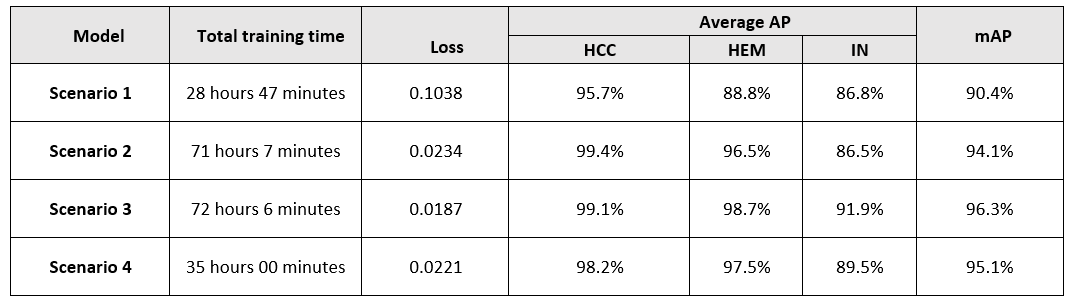
Through the process of training and testing, changing the training parameters to suit each proposed scenario, conducting comparison and evaluation, the criteria for evaluating the model are summarized in Table 1.
Conclusions
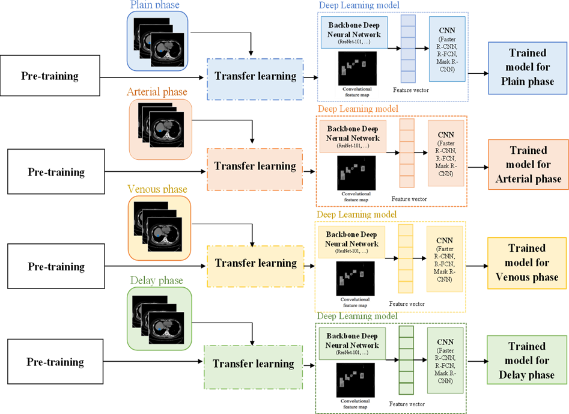
The project proposed eight scenarios and achieved notable results. First, a CT image dataset of the liver was collected before and after contrast injection to create a comprehensive input dataset. Second, the HU index, a critical medical imaging indicator, was used to enhance feature learning and create a reliable dataset with expert-annotated labels based on HU intensity variations. Third, the study employed deep learning techniques—Mask R-CNN Inception-V2, R-FCN ResNet-101, Faster R-CNN ResNet-101, and Faster R-CNN Inception-ResNet-V2—with optimized training parameters to evaluate and compare models for classifying common liver lesions, providing a basis for selecting an effective implementation model. Fourth, a four-phase classification model, incorporating CT imaging before and after the administration of Plain, Arterial, Venous, and Delay contrast agents, was proposed to enhance classification accuracy. Finally, the study compared its experimental model with recent relevant research, revealing that the Faster R-CNN ResNet-101 model achieved the highest accuracy with an average mAP of 97.4%, indicating that this model is a promising and effective option for detecting and classifying liver lesions, supporting doctors in patient care.
References
- Abadi, Martín et al. 2016. “TensorFlow: A System for Large-Scale Machine Learning.”
- Abdel-Misih, Sherif R. Z., and Mark Bloomston. 2010. “Liver Anatomy.” Surgical Clinics of North America 90(4): 643–53.
- Adam, Andreas, and Adrian K. Dixon. 2014. Grainger & Allison’s Diagnostic Radiology. 6th ed. Churchill Livingstone.
- Boureau, Y-Lan, Francis Bach, Yann LeCun, and Jean Ponce. 2010. “Learning Mid-Level Features for Recognition.” In 2010 IEEE Computer Society Conference on Computer Vision and Pattern Recognition, IEEE, 2559–66.
- Bran, William E. 2012. Fundamentals of Diagnostic Radiology. 4th ed. Lippincott Williams & Wilkin.